Page 165 - EJMO-9-2
P. 165
Eurasian Journal of
Medicine and Oncology Genetic insights into CAD drug targets
AA
B
C
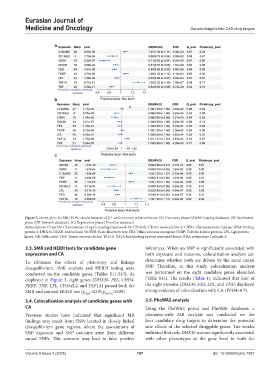
Figure 2. Forest plots for MR (IVW) results between eQTL and coronary atherosclerosis. (A) Discovery phase (GWAS Catalog database). (B) Replication
phase (UK Biobank database). (C) Replication phase (FinnGen database)
Abbreviations: C12orf39: Chromosome 12 open-reading framework 39; CD164l2: CD164 molecule like 2; CHD4: Chromodomain helicase DNA binding
protein 4; DHX36: DEAH-box helicase 36; FDR: False discovery rate; FES: Feline sarcoma oncogene; FKRP: Fukutin related protein; LPL: Lipoprotein
lipase; OR: Odds ratio; TNF: Tumor necrosis factor; TAF1A: TATA-box binding protein associated factor, RNA polymerase I subunit A.
3.3. SMR and HEIDI tests for candidate gene inferences. When an SNP is significantly associated with
expression and CA both exposure and outcome, colocalization analysis can
To eliminate the effects of pleiotropy and linkage determine whether both are driven by the same causal
disequilibrium, SMR analysis and HEIDI testing were SNP. Therefore, in this study, colocalization analysis
conducted on the candidate genes (Tables S11-S13). As was performed on the eight candidate genes identified
displayed in Figure 3, eight genes (DHX36, FES, CHD4, (Table S14). The results (Table 1) indicated that four of
FKRP, TNF, LPL, CD164L2, and TAF1A) passed both the the eight proteins (DHX36, FES, LPL, and TNF) displayed
SMR analysis and HEIDI test (p SMR <0.05; p HEIDI >0.05). strong evidence of colocalization with CA (PPH4>0.7).
3.4. Colocalization analysis of candidate genes with 3.5. PheWAS analysis
CA Using the PheWAS portal and PheWeb databases, a
Previous studies have indicated that significant MR phenome-wide MR analysis was conducted on the
findings may result from SNPs located in closely linked four candidate drug targets to determine the potential
disequilibrium gene regions, where the associations of side effects of the selected druggable genes. The results
SNP exposure and SNP outcome arise from different indicated that only DHX36 was not significantly associated
causal SNPs. This scenario may lead to false positive with other phenotypes at the gene level in both the
Volume 9 Issue 2 (2025) 157 doi: 10.36922/ejmo.7387