Page 166 - EJMO-9-2
P. 166
Eurasian Journal of
Medicine and Oncology Genetic insights into CAD drug targets
A B C
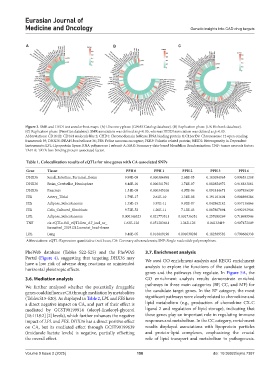
Figure 3. SMR and HEDI test annular heat maps. (A) Discovery phase (GWAS Catalog database). (B) Replication phase (UK Biobank database).
(C) Replication phase (FinnGen database). SMR association was defined as p<0.05, whereas HEIDI association was defined as p>0.05.
Abbreviations: CD164l2: CD164 molecule like 2; CHD4: Chromodomain helicase DNA binding protein 4; C12orf39: Chromosome 12 open-reading
framework 39; DHX36: DEAH-box helicase 36; FES: Feline sarcoma oncogene; FKRP: Fukutin related protein; HEIDI: Heterogeneity in Dependent
Instruments; LPL: Lipoprotein lipase; RNA polymerase I subunit A; SMR: Summary-data-based Mendelian Randomization; TNF: tumor necrosis factor;
TAF1A: TATA-box binding protein associated factor.
Table 1. Colocalization results of eQTLs for nine genes with CA‑associated SNPs
Gene Tissue PPH 0 PPH 1 PPH 2 PPH 3 PPH 4
DHX36 Small_Intestine_Terminal_Ileum 9.99E-08 0.000386898 2.68E-05 0.103054965 0.896531208
DHX36 Brain_Cerebellar_Hemisphere 6.46E-10 0.000311792 1.74E-07 0.082854972 0.916833061
DHX36 Pancreas 1.51E-08 0.000345106 4.07E-06 0.091814671 0.907836139
FES Artery_Tibial 1.79E-17 2.61E-10 1.31E-08 0.191101601 0.808898386
FES Adipose_Subcutaneous 1.35E-15 5.97E-11 9.92E-07 0.042862142 0.957136866
FES Cells_Cultured_fibroblasts 9.72E-52 1.20E-11 7.13E-43 0.007807094 0.992192906
LPL Adipose_Subcutaneous 0.000146323 0.021773511 0.001736351 0.257658269 0.718685546
TNF cis-eQTLs-full_eQTLGen_AF_incl_nr_ 1.63E-126 0.051203661 1.34E-126 0.04132419 0.907472149
formatted_20191212.new.txt_besd-dense
LPL Lung 9.46E-05 0.016819198 0.001030288 0.182389582 0.799666338
Abbreviations: eQTL: Expression quantitative trait locus; CA: Coronary atherosclerosis; SNP: Single nucleotide polymorphism.
PheWeb database (Tables S22-S25) and the PheWAS 3.7. Enrichment analysis
Portal (Figure 4), suggesting that targeting DHX36 may We used GO enrichment analysis and KEGG enrichment
have a low risk of adverse drug reactions or unintended analysis to explore the functions of the candidate target
horizontal pleiotropic effects.
genes and the pathways they regulate. In Figure 5A, the
3.6. Mediation analysis GO enrichment analysis results demonstrate enriched
We further analyzed whether the potentially druggable pathways in three main categories (BP, CC, and MF) for
genes could influence CA through mediation by metabolites the candidate target genes. In the BP category, the most
(Tables S15-S20). As displayed in Table 2, LPL and FES have significant pathways were closely related to chemokine and
a direct negative impact on CA, and part of their effect is lipid metabolism (e.g., production of chemokine CX-C
mediated by GCST90199914 (oleoyl-linoleoyl-glycerol ligand 2 and regulation of lipid storage), indicating that
[18:1/18:2] [2] levels), which further enhances the negative these genes play an important role in regulating immune
impact of LPL and FES. DHX36 has a direct positive effect responses and metabolism. In the CC category, enrichment
on CA, but its mediated effect through GCST90199639 results displayed associations with lipoprotein particles
(imidazole lactate levels) is negative, partially offsetting and protein-lipid complexes, emphasizing the crucial
the overall effect. role of lipid transport and metabolism in pathogenesis.
Volume 9 Issue 2 (2025) 158 doi: 10.36922/ejmo.7387