Page 19 - GPD-2-3
P. 19
Gene & Protein in Disease Signatures construction strategies for TC
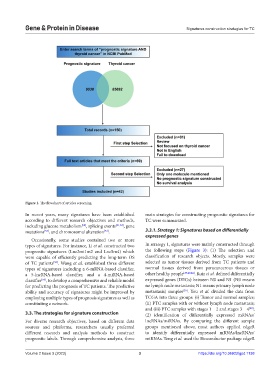
Figure 1. The flowchart of articles screening.
In recent years, many signatures have been established main strategies for constructing prognostic signatures for
according to different research objectives and methods, TC were summarized.
including glucose metabolism , splicing events [51,52] , gene
[50]
mutations , and chromosomal alteration . 3.3.1. Strategy 1: Signatures based on differentially
[54]
[53]
expressed genes
Occasionally, some studies contained two or more
types of signatures. For instance, Li et al. constructed two In strategy 1, signatures were mainly constructed through
prognostic signatures (Lnc2mi1m2 and Lnc5m4) which the following steps (Figure 3): (1) The selection and
were capable of efficiently predicting the long-term OS classification of research objects. Mostly, samples were
of TC patients . Wang et al. established three different selected as tumor tissues derived from TC patients and
[55]
types of signatures including a 6-mRNA-based classifier, normal tissues derived from paracancerous tissues or
a 5-lncRNA-based classifier, and a 4-miRNA-based other healthy people [29,40,44] . Ruiz et al. defined differentially
classifier , to develop a comprehensive and reliable model expressed genes (DEGs) between N0 and N1 (N0 means
[56]
for predicting the prognosis of TC patients. The predictive no lymph node metastasis; N1 means primary lymph node
[20]
ability and accuracy of signatures might be improved by metastasis) samples . You et al. divided the data from
employing multiple types of prognosis signatures as well as TCGA into three groups: (i) Tumor and normal samples;
constituting a network. (ii) PTC samples with or without lymph node metastasis;
and (iii) PTC samples with stages 1 – 2 and stages 3 – 4 ;
[33]
3.3. The strategies for signature construction (2) identification of differentially expressed mRNAs/
For diverse research objectives, based on different data lncRNAs/miRNAs. By comparing the different sample
sources and platforms, researchers usually preferred groups mentioned above, most authors applied edgeR
different research and analysis methods to construct to identify differentially expressed mRNAs/lncRNAs/
prognostic labels. Through comprehensive analysis, three miRNAs. Teng et al. used the Bioconductor package edgeR
Volume 2 Issue 3 (2023) 3 https://doi.org/10.36922/gpd.1138