Page 60 - GPD-2-3
P. 60
Gene & Protein in Disease SARS-CoV-2 Omicron variants in Iraq
significant . On the other hands, N679K and P681H
[54]
mutations have been found to regulate the fusogenic ability
of Omicron variants .
[55]
Q954H and N969K mutations, which are located in the
HR domain, have been found in all Omicron sub-lineages,
and only affect viral fusion and infection ability, since these
[56]
mutations do not impact the HR1-HR2 connections .
5. Conclusion
This study analyzed all the available data about the
diversity of SARS-CoV-2 Omicron sub-lineages detected
in Iraq. The BA.1, BA.1.1, BA.2, and BA.5.1 variants are
among the Omicron sub-lineages commonly found in
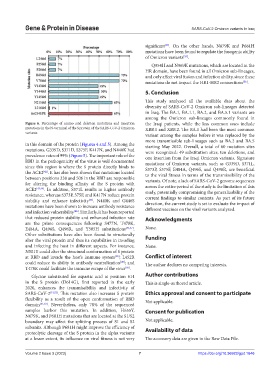
Figure 6. Percentage of amino acid deletion mutations and insertion the Iraqi patients, while the less common ones include
mutations in the N-terminal of the S protein of the SARS-CoV-2 Omicron XBB.1 and XBB.2. The BA.1 had been the most common
variants.
variant among the samples before it was replaced by the
more transmissible sub-lineages such as BA.2 and BA.5
in this domain of the protein (Figures 4 and 5). Among the starting May 2022. Overall, a total of 60 mutation sites
mutations, G339D, S371D, S375F, K417N, and N440K had were recognized: 49 substitution sites, ten deletions, and
prevalence rates of 99% (Figure 5). The important role of the one insertion from the Iraqi Omicron variants. Signature
RBD in the pathogenicity of the virus is well documented mutations of Omicron variants, such as G339D, S371L,
since this region is where the S protein directly binds to S373P, S375F, E484A, Q496S, and Q498R, are beneficial
the ACE2 . It has also been shown that mutations located to the viral fitness in terms of the transmissibility of the
[42]
between positions 338 and 506 in the RBD are responsible variants. Of note, a lack of SARS-CoV-2 genome sequences
for altering the binding affinity of the S protein with across the entire period of the study is the limitation of this
ACE2 [43,44] . In addition, S371L results in higher antibody study, potentially compromising the generalizability of the
resistance, whereas S373P, 375F, and K417N reduce protein current findings to similar contexts. As part of its future
stability and enhance infectivity . N440K and G446S direction, the current study is set to evaluate the impact of
[45]
mutations have been shown to increase antibody resistance different vaccines on the viral variants analyzed.
and infection vulnerability . Similarly, it has been reported
[46]
that reduced protein stability and enhanced infection rate Acknowledgments
are the prime consequences following S477N, T478K,
E484A, Q496S, Q498R, and Y505H substitutions [45,47] . None.
Other substitutions have also been found to structurally
alter the viral protein and thus its capabilities in invading Funding
and infecting the host in different aspects. For instance, None.
N501Y could alter the structural conformation of S protein
at RBD and invade the host’s immune system ; L452R Conflict of interest
[48]
could reduce its ability in antibody neutralization ; and The author declares no competing interests.
[49]
T478K could facilitate the immune escape of the virus .
[50]
Glycine substituted for aspartic acid at position 614 Author contributions
in the S protein (D614G), first reported in the early This is single-authored article.
2020, enhances the transmissibility and infectivity of
SARS-CoV-2 [51,52] . This mutation also increases S protein Ethics approval and consent to participate
flexibility as a result of the open conformation of RBD Not applicable.
domain [51,53] . Nevertheless, only 78% of the sequenced
samples harbor this mutation. In addition, H655Y, Consent for publication
N679K, and P681H mutations that are located at the S1/S2
boundary may affect the splitting process of S1 and S2 Not applicable.
subunits. Although P681H might improve the efficiency of Availability of data
proteolytic cleavage of the S protein in the alpha variants
at a lesser extent, its influence on viral fitness is not very The accessory data are given in the Raw Data File.
Volume 2 Issue 3 (2023) 7 https://doi.org/10.36922/gpd.1646