Page 56 - GPD-2-3
P. 56
Gene & Protein in Disease SARS-CoV-2 Omicron variants in Iraq
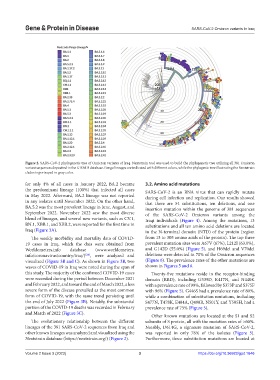
Figure 2. SARS-CoV-2 phylogenetic tree of Omicron variants of Iraq. Nextstrain tool was used to build the phylogenetic tree utilizing all 381 Omicron
variant sequences deposited in the GISAID database. Pango lineages are indicated with different colors, while the phylogenic tree illustrating the Nextstrain
clades is portrayed in gray color.
for only 4% of all cases in January 2022, BA.2 became 3.2. Amino acid mutations
the predominant lineage (100%) that infected all cases SARS-CoV-2 is an RNA virus that can rapidly mutate
in May 2022. Afterward, BA.2 lineage was not reported during cell infection and replication. Our results showed
in any isolates until November 2022. On the other hand, that there are 54 substitutions, ten deletions, and one
BA.5.2 was the most prevalent lineage in June, August, and insertion mutation within the genome of 381 sequences
September 2022. November 2022 saw the most diverse of the SARS-CoV-2 Omicron variants among the
blend of lineages, and several new variants, such as CV.1, Iraqi individuals (Figure 4). Among the mutations, 13
BN.1, XBB.1, and XBB.2, were reported for the first time in substitutions and all ten amino acid deletions are located
Iraq (Figure 3A). in the N-terminal domain (NTD) of the protein (region
The weekly morbidity and mortality data of COVID- from 13 to 305 amino acids of the protein). The top three
19 cases in Iraq, which the data were obtained from prevalent mutation sites were A67V (67%), L212I (63.0%),
Worldometers.info database (www.worldometers. and G142D (55.0%) (Figure 5), and H69del and V70del
info/coronavirus/country/iraq/) , were analyzed and deletions were detected in 70% of the Omicron sequences
[24]
visualized (Figure 3B and C). As shown in Figure 3B, two (Figure 6). The prevalence rates of the other mutations are
waves of COVID-19 in Iraq were noted during the span of shown in Figures 5 and 6.
this study. The majority of the confirmed COVID-19 cases Twenty-five mutations reside in the receptor-binding
were recorded during the period between December 2021 domain (RBD), including G339D, K417N, and N440K
and February 2022, and toward the end of March 2022, a less with a prevalence rate of 99%, followed by S373P and S375F
severe form of the disease prevailed as the most common with 96% (Figure 5). G446S had a prevalence rate of 80%,
form of COVID-19, with the same trend persisting until while a combination of substitution mutations, including
the end of July 2022 (Figure 3B). Notably, the substantial S477N, T478K, E484A, Q498R, N501Y, and Y505H, had a
portion of the COVID-19 deaths was recorded in February prevalence rate of 75% (Figure 5).
and March of 2022 (Figure 3C).
Other known mutations are located at the S1 and S2
The evolutionary relationship between the different subunits of S protein, all with the mutation rates of >60%.
lineages of the 381 SARS-CoV-2 sequences from Iraq and Notably, D614G, a signature mutation of SARS-CoV-2,
other known lineages was analyzed and visualized using the was reported in only 78% of the isolates (Figure 5).
Nextstrain database (https://nextstrain.org/) (Figure 2). Furthermore, three substitution mutations are located at
Volume 2 Issue 3 (2023) 3 https://doi.org/10.36922/gpd.1646