Page 50 - GPD-3-1
P. 50
Gene & Protein in Disease Prognostic potential of LMNB2 in LPS
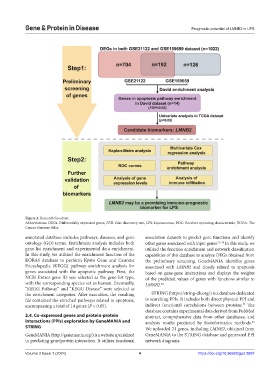
Figure 2. Research flowchart.
Abbreviations: DEGs: Differentially expressed genes; FDR: false discovery rate; LPS: Liposarcoma; ROC: Receiver operating characteristic; TCGA: The
Cancer Genome Atlas.
annotated database includes pathways, diseases, and gene association datasets to predict gene functions and identify
ontology (GO) terms. Enrichment analysis includes both other genes associated with input genes. 32-34 In this study, we
gene-list enrichment and experimental data enrichment. utilized the function enrichment and network classification
In this study, we utilized the enrichment function of the capabilities of this database to analyze DEGs obtained from
KOBAS database to perform Kyoto Gene and Genome the preliminary screening. GeneMANIA identifies genes
Encyclopedia (KEGG) pathway enrichment analysis for associated with LMNB2 and closely related to apoptosis
genes associated with the apoptotic pathway. First, the based on gene-gene interactions and displays the weights
NCBI Entrez gene ID was selected as the gene-list type, of the predicted values of genes with functions similar to
with the corresponding species set as human. Eventually, LMNB2. 34
“KEGG Pathway” and “KEGG Disease” were selected as
the enrichment categories. After execution, the resulting STRING (https://string-db.org/) is a database dedicated
file contained the enriched pathways related to apoptosis, to searching PPIs. It includes both direct physical PPI and
35
encompassing a total of 14 genes (P < 0.05). indirect functional correlations between proteins. The
database contains experimental data derived from PubMed
2.4. Co-expressed genes and protein-protein abstract, comprehensive data from other databases, and
interactions (PPIs) exploration by GeneMANIA and analysis results predicted by bioinformatics methods.
36
STRING We uploaded 21 genes, including LMNB2, obtained from
GeneMANIA (http://genemania.org/) is a website specialized GeneMANIA to the STRING database and generated PPI
in predicting gene/protein interaction. It utilizes functional network diagrams.
Volume 3 Issue 1 (2024) 4 https://doi.org/10.36922/gpd.2607