Page 154 - GPD-4-1
P. 154
Gene & Protein in Disease FXR1 modulates gene expression in cancer
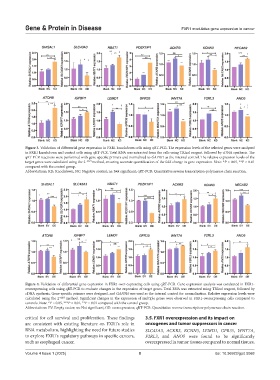
Figure 3. Validation of differential gene expression in FXR1 knockdown cells using qRT-PCR. The expression levels of the selected genes were analyzed
in FXR1 knockdown and control cells using qRT-PCR. Total RNA was extracted from the cells using TRIzol reagent, followed by cDNA synthesis. The
qRT-PCR reactions were performed with gene-specific primers and normalized to GAPDH as the internal control. The relative expression levels of the
target genes were calculated using the 2 −ΔΔCt method, ensuring accurate quantification of the fold change in gene expression. Note: *P < 0.05, **P < 0.01
compared with the control group.
Abbreviations: KD: Knockdown; NC: Negative control; ns: Not significant; qRT-PCR: Quantitative reverse transcription-polymerase chain reaction.
Figure 4. Validation of differential gene expression in FXR1 over-expressing cells using qRT-PCR. Gene expression analysis was conducted in FXR1-
overexpressing cells using qRT-PCR to evaluate changes in the expression of target genes. Total RNA was extracted using TRIzol reagent, followed by
cDNA synthesis. Gene-specific primers were designed, and GAPDH was used as the internal control for normalization. Relative expression levels were
calculated using the 2 −ΔΔCt method. Significant changes in the expression of multiple genes were observed in FXR1-overexpressing cells compared to
controls. Note: *P < 0.05, ***P < 0.01, **P < 0.01 compared with the control group.
Abbreviations: EV: Empty vector; ns: Not significant; OE: overexpression; qRT-PCR: Quantitative reverse transcription-polymerase chain reaction.
critical for cell survival and proliferation. These findings 3.5. FXR1 overexpression and its impact on
are consistent with existing literature on FXR1’s role in oncogenes and tumor suppressors in cancer
RNA metabolism, highlighting the need for future studies SLC43A3, ACKR3, KCNN3, LEMD1, GPR35, WNT7A,
to explore FXR1’s regulatory pathways in specific cancers, F2RL3, and ANO5 were found to be significantly
such as esophageal cancer. overexpressed in tumor tissues compared to normal tissues,
Volume 4 Issue 1 (2025) 8 doi: 10.36922/gpd.5068