Page 48 - GTM-1-1
P. 48
Global Translational Medicine Bioinformatics analysis of dilated cardiomyopathy
4. Discussion
The proteins encoded by the genes of DCM have a wide
range of biological functions; therefore, there are a
diverse array of mechanisms underlying the pathogenesis
of DCM, including mutations in cytoskeletal proteins,
mitochondrial proteins, and bridging proteins . DCM
[20]
with an inherited trait is called familial DCM and accounts
[21]
for approximately 20 – 50% of patients with DCM .
Autosomal dominant inheritance usually occurs in the
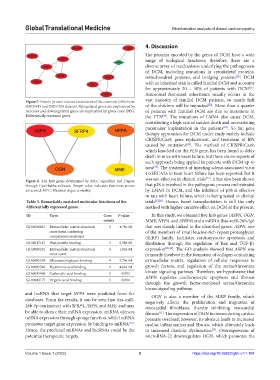
Figure 5. Protein-protein interaction network of the common DEGs from vast majority of familial DCM patients, so nearly half
[22]
GSE29819 and GSE57338 datasets. Upregulated genes are emphasized by of the children will be impacted . More than a quarter
red color and downregulated genes are emphasized by green color. DEG: of patients with familial DCM are due to mutations in
Differentially expressed genes. the TTN . The mutations of LMNA also cause DCM,
[23]
contributing a high rate of sudden death and necessitating
pacemaker implantation in the patients . So far, gene
[24]
therapy approaches for DCM under study mainly include
CRISPR/Cas9, gene replacement, and treatment of BPs
[25]
caused by mutations . The method of CRISPR/Cas9,
which knocked out the PLN gene, has been found to delay
death in mice with heart failure, but there are no reports of
such approach being applied in patients with DCM up to
[26]
now . The treatment of injecting adeno-associated virus
1/SERCA2a to treat heart failure has been reported, but it
[27]
Figure 6. The hub genes determined by MCC algorithm and Degree was not effective in clinical trials . It has also been shown
through CytoHubba software. Deeper color indicates that more points that p38 is involved in the pathogenic process orchestrated
are scored. MCC: Maximal clique centrality. by LMNA in DCM, and the inhibitor of p38 is effective
in rats with heart failure, which is being tested in clinical
Table 3. Remarkably enriched molecular functions of the trials [25,28] . Hence, heart transplantation is still the only
differentially expressed genes. method with higher curative effect on DCM at the present.
ID Term Gene P value In this study, we obtained five hub genes (ASPN, OGN,
counts MME, NPPA, and SFRP4) and a miRNA (hsa-miR-26b-5p)
GO:0030021 Extracellular matrix structural 3 4.78e-06 that was closely linked to the identified genes. ASPN, one
constituent conferring of the members of small leucine-rich repeat proteoglycan
compression resistance (SLRP) family, facilitates cardiomyocyte apoptosis and
GO:0017147 Wnt-protein binding 3 2.38e-05 fibrillation through the regulation of Bax and TGF-β1
GO:0005201 Extracellular matrix structural 4 1.03e-04 expression [29,30] . The GO analysis showed that ASPN was
constituent primarily involved in the formation of collagen-containing
GO:0005539 Glycosaminoglycan binding 4 3.79e-04 extracellular matrix, regulation of cellular responses to
GO:0005540 Hyaluronic acid binding 2 4.62e-04 growth factors, and regulation of the serine/threonine
GO:0031406 Carboxylic acid binding 3 0.003 kinase signaling pathway. Therefore, we hypothesize that
GO:0043177 Organic acid binding 3 0.004 ASPN regulates cardiomyocyte apoptosis and fibrosis
through the growth factor-mediated serine/threonine
kinase signaling pathway.
and lncRNA that target NPPA were predicted from the
OGN is also a member of the SLRP family, which
databases. From the results, it can be seen that hsa-miR- negatively affects the proliferation and migration of
26b-5p can interact with SFRP4, ASPN, and MEE, and may myocardial fibroblasts, thereby inhibiting myocardial
be able to silence their mRNA expression. miRNA silences fibrosis . The expression of OGN increases during cardiac
[31]
mRNA expression through sponge function, while lncRNA pressure overload; however, its absence leads to increased
promotes target gene expression by binding to miRNA . cardiac inflammation and fibrosis, which ultimately leads
[13]
Hence, the predicted miRNAs and lncRNAs could be the to increased diastolic dysfunction . Overexpression of
[32]
potential therapeutic targets. microRNA-22 downregulates OGN, which promotes the
Volume 1 Issue 1 (2022) 8 https://doi.org/10.36922/gtm.v1i1.104