Page 155 - IJB-10-1
P. 155
International Journal of Bioprinting 3D-printed micro-perfused culture device
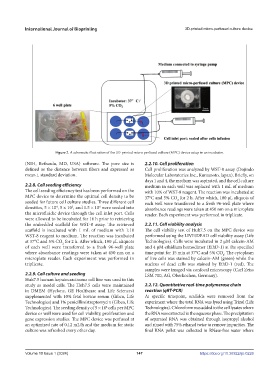
Figure 2. A schematic illustration of the 3D-printed micro-perfused culture (MPC) device setup in an incubator.
(NIH, Bethesda, MD, USA) software. The pore size is 2.2.10. Cell proliferation
defined as the distance between fibers and expressed as Cell proliferation was analyzed by WST-8 assay (Dojindo
mean ± standard deviation. Molecular Laboratories Inc., Kumanoto, Japan). Briefly, on
days 1 and 4, the medium was aspirated, and the cell culture
2.2.8. Cell seeding efficiency medium in each well was replaced with 1 mL of medium
The cell seeding efficiency test has been performed on the with 10% of WST-8 reagent. The reaction was incubated at
MPC device to determine the optimal cell density to be 37°C and 5% CO for 2 h. After which, 100 µL aliquots of
2
seeded for future cell culture studies. Three different cell each well were transferred to a fresh 96-well plate where
densities, 5 × 10 , 5 × 10 , and 1.5 × 10 were seeded into absorbance readings were taken at 450 nm on a microplate
4
6
5
the microfluidic device through the cell inlet port. Cells reader. Each experiment was performed in triplicate.
were allowed to be incubated for 16 h prior to retrieving
the embedded scaffold for WST-8 assay. The retrieved 2.2.11. Cell viability analysis
scaffold is incubated with 1 mL of medium with 1:10 The cell viability test of Huh7.5 on the MPC device was
WST-8 reagent to medium. The reaction was incubated performed using the LIVE/DEAD cell viability assay (Life
at 37°C and 5% CO for 2 h. After which, 100 µL aliquots Technologies). Cells were incubated in 2 µM calcein-AM
2
of each well were transferred to a fresh 96-well plate and 4 µM ethidium homodimer (EthD-1) at the specified
where absorbance readings were taken at 450 nm on a time point for 15 min at 37°C and 5% CO . The cytoplasm
2
microplate reader. Each experiment was performed in of live cells was stained by calcein-AM (green) while the
triplicate. nucleus of dead cells was stained by EthD-1 (red). The
samples were imaged via confocal microscopy (Carl Zeiss
2.2.9. Cell culture and seeding LSM 700, AG, Oberkochen, Germany).
Huh7.5 human hepatocarcinoma cell line was used in this
study as model cells. The Huh7.5 cells were maintained 2.2.12. Quantitative real-time polymerase chain
in DMEM (Hyclone, GE Healthcare and Life Sciences) reaction (qRT-PCR)
supplemented with 10% fetal bovine serum (Gibco, Life At specific timepoint, scaffolds were removed from the
Technologies) and 1% penicillin/streptomycin (Gibco, Life experiment where the total RNA was lysed using Trizol (Life
Technologies). The seeding density of 5 × 10 cells per MPC Technologies). Chloroform was added to the cell lysates where
4
device or well were used for cell viability, proliferation and the RNA was extracted in the aqueous phase. The precipitation
gene expression studies. The MPC device was perfused at of separated RNA was obtained through isopropyl alcohol
an optimized rate of 0.12 mL/h and the medium for static and rinsed with 75% ethanol twice to remove impurities. The
culture was refreshed every other day. final RNA pellet was collected in RNase-free water where
Volume 10 Issue 1 (2024) 147 https://doi.org/10.36922/ijb.0226