Page 572 - IJB-10-5
P. 572
International Journal of Bioprinting ML-generated GelMA compression database
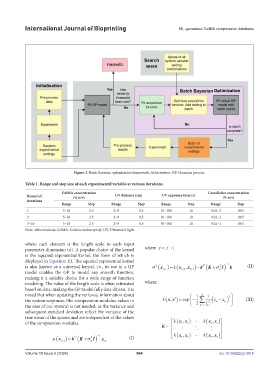
Figure 3. Batch Bayesian optimization framework. Abbreviation: GP: Gaussian process.
Table 1. Range and step size of each experimental variable at various iterations.
GelMA concentration Crosslinker concentration
Round of (% w/v) UV distance (cm) UV exposure time (s) (% w/v)
iterations
Range Step Range Step Range Step Range Step
1 5–10 2.5 2–8 0.5 10–180 10 0.01–2 0.05
2 5–10 2.5 2–8 0.5 10–180 10 0.01–1 0.05
3–10 5–10 2.5 2–8 0.5 30–180 10 0.01–1 0.05
Note: Abbreviations: GelMA: Gelatin methacryloyl; UV: Ultraviolet light.
where each element is the length scale in each input
parameter dimension (n). A popular choice of the kernel where y f
is the squared exponential kernel, the form of which is
displayed in Equation III. The squared exponential kernel
T
2
2
is also known as a universal kernel, i.e., its use in a GP x t 1 x , x t 1 K I 1 k (II)
k
k
t
n
1
model enables the GP to model any smooth function,
making it a suitable choice for a wide range of function
modeling. The value of the length scale is often estimated where:
based on data, making the GP model fully data-driven. It is
noted that when updating the variance, information about 1 4 1 2
n
'
x
the system responses (the compression modulus values in k xx, exp 2 l 2 x (III)
n
the case of our system) is not needed, as the variance and n1 n
subsequent standard deviation reflect the variance of the
true mean of the system and are independent of the values
1
1
of the compression modulus. k xx, k xx,
t
1
K =
t
t
t
x t k K I 1 y t 1: (I) k xx, k xx,
T
2
1
1
n
Volume 10 Issue 5 (2024) 564 doi: 10.36922/ijb.3814