Page 563 - IJB-9-6
P. 563
International Journal of Bioprinting Osteogenic, antibacterial CpTi-MgOCu implants
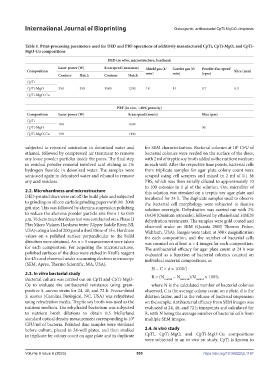
Table 1. Print-processing parameters used for DED and PBF operations of additively manufactured CpTi, CpTi-MgO, and CpTi-
MgO-Cu compositions
DED (in vitro, microstructure, hardness)
Laser power (W) Scan speed (mm/min) Shield gas (ℓ/ Carrier gas (ℓ/ Powder disc speed
Composition min) min) (rpm) Slice (mm)
Contour Hatch Contour Hatch
CpTi
CpTi-MgO 350 350 1500 1200 18 14 0.7 0.3
CpTi-MgO-Cu
PBF (in vivo, ~40% porosity)
Composition Laser power (W) Scan speed (mm/s) Slice (μm)
CpTi
180 1600
CpTi-MgO 30
CpTi-MgO-Cu 198 1440
subjected to repeated sonication in deionized water and for SEM characterization. Bacterial colonies at 10 CFU of
6
ethanol, followed by compressed air treatment to remove bacterial colonies were seeded on the surface of the discs,
any loose powder particles inside the pores. The final step with 2 ml of tryptic soy broth added as the nutrient medium
in residual powder removal involved acid etching in 1% in each well. After the respective time points, bacterial cells
hydrogen fluoride in deionized water. The samples were from triplicate samples for agar plate colony count were
sonicated again in deionized water and ethanol to remove scraped using cell scrapers and mixed in 2 ml of 0.1 M
any acid residues. PBS, which was then serially diluted to approximately 10
to 100 colonies in 1 µl of the solution. One microliter of
2.2. Microhardness and microstructure this solution was streaked on a tryptic soy agar plate and
DED-printed discs were cut off the build plate and subjected incubated for 24 h. The duplicate samples used to observe
to grinding on silicon carbide grinding papers with 80–2000 the bacterial cell morphology were subjected to fixative
grit size. This was followed by alumina suspension polishing solution overnight. Dehydration was carried out with 2%
to reduce the alumina powder particle size from 1 to 0.05 OsO4 (Osmium tetroxide), followed by ethanol and HMDS
µm. Vickers microhardness test was conducted on a Phase II dehydration treatments. The samples were gold coated and
Plus Micro Vickers Hardness tester (Upper Saddle River, NJ, observed under an SEM (Quanta 200F, Thermo Fisher,
USA) using a load of 200 g and a dwell time of 15 s. Hardness Waltham, USA). Images were taken at 300× magnification
values on a polished surface perpendicular to the build for each composition, and the number of bacterial cells
direction were obtained. An n = 5 measurement were taken was counted on at least n = 4 images for each composition.
for each composition. For acquiring the microstructures, The antibacterial efficacy for agar plate count at 24 h was
polished surfaces of the discs were etched in Kroll’s reagent evaluated as a function of bacterial colonies counted on
for 45 s and observed under a scanning electron microscope individual material compositions, as
(SEM; Apreo, Thermo Scientific, MA, USA).
N = C × d × 1000/l
2.3. In vitro bacterial study
Bacterial culture was carried out on CpTi and CpTi-MgO- R = (N control − N material )/N control × 100%
Cu to evaluate the antibacterial resistance using gram- where N is the calculated number of bacterial colonies
positive S. aureus strain for 24, 48, and 72 h. Freeze-dried observed, C is the average colony count on a plate, d is the
S. aureus (Carolina Biological, NC, USA) was rehydrated dilution factor, and l is the volume of bacterial suspension
using rehydration media. Tryptic soy broth was used as the on the sample. Antibacterial efficacy from SEM images was
nutrient medium. The rehydrated bacterium was subjected evaluated at 24, 48, and 72 h timepoints and calculated for
to nutrient broth dilutions to obtain 0.5 McFarland R, with N being the average number of bacterial cells from
standard optical density measurement corresponding to 10 8 multiple SEM images.
CFU/ml of bacteria. Polished disc samples were sterilized
before culture, placed in 24-well plates, and then studied 2.4. In vivo study
in triplicate for colony count on agar plate and in duplicate CpTi, CpTi-MgO, and CpTi-MgO-Cu compositions
were subjected to an in vivo rat study. CpTi is known to
Volume 9 Issue 6 (2023) 555 https://doi.org/10.36922/ijb.1167