Page 68 - ITPS-7-1
P. 68
INNOSC Theranostics and
Pharmacological Sciences Docking study of quinoline-3-carbaldehyde derives
A A
B
B
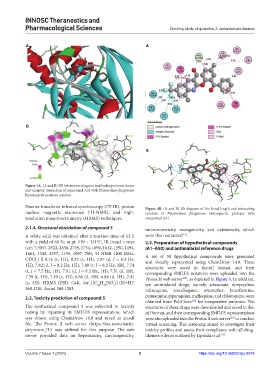
Figure 4A. (A and B) 3D interaction diagram and hydrogen bond donor
and acceptor interaction of compound A31 with Plasmodium falciparum
histoaspartic protease residues.
Fourier transform-infrared spectroscopy (FT-IR), proton Figure 4B. (A and B) 2D diagram of the bond length and interacting
nuclear magnetic resonance ( H-NMR), and high- residues of Plasmodium falciparum histoaspartic protease with
1
resolution mass spectrometry (HRMS) techniques. compound A31.
2.1.4. Structural elucidation of compound 5 immunotoxicity, mutagenicity, and cytotoxicity, which
[13]
A white solid was obtained after a reaction time of 12 h were then extracted .
with a yield of 60 %; m.pt: 119 – 121°C; IR (neat) v max 2.3. Preparation of hypothetical compounds
(cm ) 3057, 2922, 2856, 2739, 1754, 1690, 1612, 1590, 1494, (A1–A50) and antimalarial reference drugs
-1
1461, 1343, 1257, 1199, 1097, 760; H-NMR (400 MHz, A set of 50 hypothetical compounds were generated
1
CDCl ) δ 9.75 (s, 1H), 8.32 (s, 1H), 7.97 (d, J = 8.3 Hz, and visually represented using ChemDraw 14.0. These
3
1H), 7.82 (d, J = 8.2 Hz, 1H), 7.80 (t, J = 8.2 Hz, 1H), 7.74 structures were saved in the.sdf format, and their
(t, J = 7.5 Hz, 1H), 7.61 (d, J = 8.2 Hz, 1H), 7.51 (d, 1H), corresponding SMILES notations were uploaded into the
7.39 (t, 1H), 7.19 (s, 1H), 6.86 (d, 1H), 6.84 (d, 1H), 2.41 Protox II web server , as depicted in Figure 4. In addition,
[15]
(s, 3H). HRMS (ESI): Calc. for [(C H NO )] (M+H) ten antimalarial drugs, namely, artesunate, doxycycline,
+
24
17
3
368.1281, found 368.1283. tafenoquine, amodiaquine, artemether, lumefantrine,
2.2. Toxicity prediction of compound 5 primaquine, piperaquine, mefloquine, and chloroquine, were
obtained from PubChem for comparative purposes. The
[16]
The synthesized compound 5 was subjected to toxicity structures of these drugs were downloaded and saved in the.
testing by inputting its SMILES representation, which sdf format, and their corresponding SMILES representations
was drawn using ChemDraw 14.0 and saved as an.sdf were also uploaded into the Protox II web server to conduct
[16]
file. The Protox II web server (https://tox-new.charite. virtual screening. This screening aimed to investigate their
de/protox_II/) was utilized for this purpose. The web toxicity profiles and assess their compliance with all drug-
server provided data on hepatoxicity, carcinogenicity, likeness rules as outlined by Lipinski et al. [17]
Volume 7 Issue 1 (2024) 5 https://doi.org/10.36922/itps.0976