Page 69 - ITPS-7-1
P. 69
INNOSC Theranostics and
Pharmacological Sciences Docking study of quinoline-3-carbaldehyde derives
[20]
A drug development . The SMILES representations of the
compounds were submitted to the web server, and the
resulting data were carefully extracted and comprehensively
examined.
2.7. Molecular docking study
To evaluate the inhibitory potential of synthesized
compound 5, as well as the selected hypothetical
compounds, docking simulations were performed against
HAP using the PyRx 0.8 AutoDock Vina Wizard. The
macromolecules were converted to Autodock format, and a
flexible ligand to rigid protein approach was employed. All
B possible binding sites on the target protein were explored
during the docking process. The docking calculations
were performed within a cubic grid of dimensions
90 × 75 × 60 centered on the protein, encompassing the
entire protein structure. This process lasted approximately
1 h. A grid spacing of 1.00 Å was utilized to generate the
grid maps using the autogrid module of AutoDock Tools.
Each ligand underwent nine independent runs to ensure
accuracy.
Based on the identified potential binding sites,
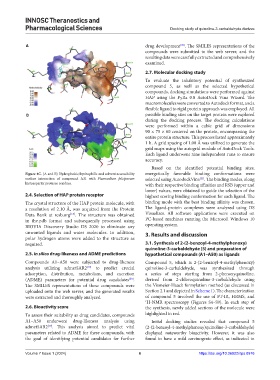
Figure 4C. (A and B) Hydrophobic/hydrophilic and solvent accessibility energetically favorable binding conformations were
surface interaction of compound A31 with Plasmodium falciparum selected using AutodockVina . The binding modes, along
[21]
histoaspartic protease residues. with their respective binding affinities and RSB (upper and
lower) values, were obtained to guide the selection of the
2.4. Selection of HAP protein receptor highest scoring binding conformation for each ligand. The
The crystal structure of the HAP protein molecule, with binding mode with the best binding affinity was chosen.
a resolution of 2.10 Å, was acquired from the Protein The ligand-protein complexes were analyzed using DS
Data Bank at rcsb.org [18] . The structure was obtained Visualizer. All software applications were executed on
in the.pdb format and subsequently processed using PC-based machines running the Microsoft Windows 10
BIOVIA Discovery Studio DS 2020 to eliminate any operating system.
unwanted ligands and water molecules. In addition, 3. Results and discussion
polar hydrogen atoms were added to the structure as
required. 3.1. Synthesis of 2-(2-benzoyl-4-methylphenoxy)
quinoline-3-carbaldehyde (5) and preparation of
2.5. In silico drug-likeness and ADME predictions hypothetical compounds (A1–A50) as ligands
Compounds A1–A50 were subjected to drug-likeness Compound 5, which is 2-(2-benzoyl-4-methylphenoxy)
analysis utilizing admetSAR2 to predict crucial quinoline-3-carbaldehyde, was synthesized through
[19]
adsorption, distribution, metabolism, and excretion a series of steps starting from 2-phenoxyquinoline,
[20]
(ADME) parameters for potential drug candidates . derived from 2-chloroquinoline-3-carbaldehyde using
The SMILES representations of these compounds were the Vismeier-Haack formylation method (as discussed in
uploaded onto the web server, and the generated results Section 2.1 and depicted in Scheme 1). The characterization
were extracted and thoroughly analyzed. of compound 5 involved the use of FT-IR, HRMS, and
1 H-NMR spectroscopy (Figures S4–S9). In each step of
2.6. Bioactivity score the synthesis, newly added sections of the molecule were
To assess their suitability as drug candidates, compounds highlighted in red.
A1–A50 underwent drug-likeness analysis using Initial docking studies revealed that compound 5
admetSAR2 . This analysis aimed to predict vital (2-(2-benzoyl-4-methylphenoxy)quinoline-3-carbaldehyde)
[19]
parameters related to ADME for these compounds, with displayed noteworthy bioactivity. However, it was also
the goal of identifying potential candidates for further found to have a mild carcinogenic effect, as indicated in
Volume 7 Issue 1 (2024) 6 https://doi.org/10.36922/itps.0976