Page 99 - ITPS-7-1
P. 99
INNOSC Theranostics and
Pharmacological Sciences Plants immunoactivity: In silico study
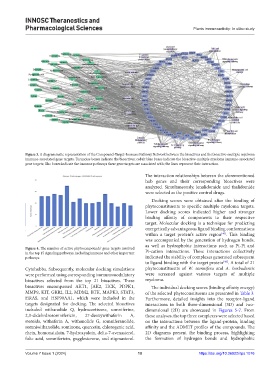
Figure 3. A diagrammatic representation of the Compound-Target-Immune Pathway Network between the bioactives and the bioactive-multiple myeloma
immune-associated gene targets. Turquoise boxes indicate the bioactives; cobalt blue boxes indicate the bioactive-multiple myeloma immune-associated
gene targets; lilac boxes indicate the immune pathways these gene targets are associated with; the lines represent their interaction.
The interaction relationships between the aforementioned
hub genes and their corresponding bioactives were
analyzed. Simultaneously, lenalidomide and thalidomide
were selected as the positive control drugs.
Docking scores were obtained after the binding of
phytoconstituents to specific multiple myeloma targets.
Lower docking scores indicated higher and stronger
binding affinity of components to their respective
target. Molecular docking is a technique for predicting
energetically advantageous ligand binding conformations
within a target protein’s active region . This binding
[40]
was accompanied by the generation of hydrogen bonds,
as well as hydrophobic interactions such as Pi-Pi and
Figure 4. The number of active phytocompounds’ gene targets involved
in the top 15 signaling pathways, including immune and other important Pi-cation interactions. These interactions collectively
pathways. indicated the stability of complexes generated subsequent
to ligand binding with the target protein . A total of 21
[40]
Cytohubba. Subsequently, molecular docking simulations phytoconstituents of W. somnifera and A. barbadensis
were performed using corresponding immunomodulatory were screened against various targets of multiple
bioactives selected from the top 21 bioactives. These myeloma.
bioactives encompassed AKT1, JAK2, HCK, PDPK1, The individual docking scores (binding affinity energy)
MMP9, KIT, GRB2, IL2, MDM2, BTK, MAPK3, STAT3, of the selected phytoconstituents are presented in Table 3.
HRAS, and HSP90AA1, which were included in the Furthermore, detailed insights into the receptor-ligand
targets designated for docking. The selected bioactives interactions in both three-dimensional (3D) and two-
included withanolide Q, hydrocortisone, somniferine, dimensional (2D) are showcased in Figures 5-7. From
2,3-didehrdrosomnifericin, 27-deoxywithaferin A, these analyses, the top three complexes were selected based
steroids, withaferin A, withanolide G, somniferanolide, on the interactions between the ligand-protein, binding
somniwithanolide, sominone, quercetin, chlorogenic acid, affinity and the ADMET profiles of the compounds. The
rhein, homonataloin, 7-hydroxyaloin, delta-7-avenasterol, 2D diagrams present the binding process, highlighting
folic acid, somnifericin, gugglesterone, and stigmasterol. the formation of hydrogen bonds and hydrophobic
Volume 7 Issue 1 (2024) 10 https://doi.org/10.36922/itps.1076