Page 14 - GPD-1-2
P. 14
Gene & Protein in Disease DNA methylation and gene expression on rats with protein malnutrition
Table 5. Sample gene transcript coverage depth statistics.
Coverage LPE_1 LPE_2 LPE_3 LPE_4 LPF_1 LPF_2 LPF_3 LPF_4 CON_1 CON_2 CON_3 CON_4
0–1 26.57% 23.46% 36.30% 27.12% 42.70% 26.98% 27.31% 26.59% 24.52% 30.72% 27.31% 20.55%
2–5 28.79% 22.83% 34.57% 27.81% 31.92% 29.48% 28.08% 27.15% 24.29% 33.42% 28.21% 17.64%
6–10 14.65% 13.23% 10.26% 14.86% 7.92% 15.03% 14.57% 15.32% 13.61% 13.32% 14.87% 9.39%
11–15 7.55% 8.32% 3.98% 7.97% 3.57% 7.24% 7.59% 7.80% 8.30% 5.69% 7.79% 6.09%
16–20 4.62% 5.65% 2.52% 4.51% 1.89% 4.27% 4.43% 4.76% 5.60% 3.18% 4.46% 4.91%
21–25 2.89% 4.31% 1.72% 3.20% 1.46% 2.89% 3.05% 3.19% 3.62% 2.27% 2.95% 4.17%
26–30 2.17% 3.16% 1.26% 2.05% 1.05% 2.06% 2.17% 2.17% 2.87% 1.60% 2.01% 3.25%
>30 12.76% 19.03% 9.39% 12.48% 9.49% 12.07% 12.80% 13.01% 17.19% 9.80% 12.41% 34.00%
LPE: Early-life low-protein group, LPF: Fetal low-protein group, CON: Control group
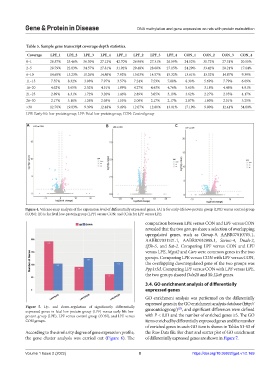
A B C
Figure 4. Volcano map analysis of the expression level of differentially expressed genes. (A) is for early-life low-protein group (LPE) versus control group
(CON); (B) is for fetal low-protein group (LPF) versus CON; and (C) is for LPF versus LPE.
comparison between LPE versus CON and LPF versus CON
revealed that the two groups share a selection of overlapping
upregulated genes, such as Gimap-9, AABR07010705.1,
AABR07031521.1, AABR07032888.1, Serinc-4, Dnah-2,
Sf3b-5, and Sat-2. Comparing LPF versus CON and LPF
versus LPE, Mgat2 and Cars were common genes in the two
groups. Comparing LPE versus CON with LPF versus CON,
the overlapping downregulated gene of the two groups was
Ppp1r3d. Comparing LPF versus CON with LPF versus LPE,
the two groups shared Ddx28 and Slc12a9 genes.
3.4. GO enrichment analysis of differentially
expressed genes
GO enrichment analysis was performed on the differentially
expressed genes in the GO enrichment analysis database (http://
Figure 5. Up- and down-regulation of significantly differentially geneontology.org/) , and significant differences were defined
[25]
expressed genes in fetal low-protein group (LPF) versus early-life low-
protein group (LPE), LPF versus control group (CON), and LPE versus with P < 0.01 and the number of enriched genes ≥5. The GO
CON groups. items enriched by differentially expressed genes and the number
of enriched genes in each GO item is shown in Tables S1-S3 of
According to the similarity degree of gene expression profile, the Raw Data file. Bar chart and scatter plot of GO enrichment
the gene cluster analysis was carried out (Figure 6). The of differentially expressed genes are shown in Figure 7.
Volume 1 Issue 2 (2022) 8 https://doi.org/10.36922/gpd.v1i2.169