Page 26 - GPD-1-2
P. 26
Gene & Protein in Disease DNA methylation and gene expression on rats with protein malnutrition
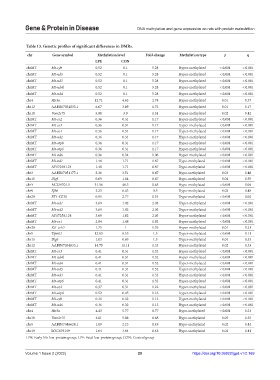
Table 13. Genetic profiles of significant differences in DMRs.
chr Gene symbol Methylation level Fold‑change Methylation type p q
LPE CON
chrMT Mt-cyb 0.52 0.1 5.28 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd5 0.52 0.1 5.28 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd3 0.52 0.1 5.28 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd4l 0.52 0.1 5.28 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd4 0.52 0.1 5.28 Hyper-methylated < 0.001 < 0.001
chr4 Htr5a 12.71 4.63 2.74 Hyper-methylated 0.01 0.37
chr12 AABR07034833.1 6.67 3.89 1.72 Hyper-methylated 0.01 0.17
chr18 Vom2r75 5.88 3.9 1.51 Hyper-methylated 0.02 0.42
chrMT Mt-co2 0.36 0.31 1.17 Hyper-methylated < 0.001 < 0.001
chrMT Mt-co3 0.36 0.31 1.17 Hyper-methylated < 0.001 < 0.001
chrMT Mt-co1 0.36 0.31 1.17 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd2 0.36 0.31 1.17 Hyper-methylated < 0.001 < 0.001
chrMT Mt-atp8 0.36 0.31 1.17 Hyper-methylated < 0.001 < 0.001
chrMT Mt-atp6 0.36 0.31 1.17 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd6 0.36 0.34 1.06 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd1 1.48 1.71 0.87 Hypo-methylated < 0.001 < 0.001
chrMT AY172581.24 1.48 1.71 0.87 Hypo-methylated < 0.001 < 0.001
chr3 AABR07051177.1 2.36 3.51 0.67 Hypo-methylated 0.03 0.48
chr15 Dlg5 0.69 1.04 0.67 Hypo-methylated 0.04 0.55
chrY AC239701.1 31.56 48.5 0.65 Hypo-methylated < 0.001 0.04
chr9 Efhb 3.23 6.43 0.5 Hypo-methylated 0.03 0.48
chr20 RT1-CE10 0.95 2.77 0.34 Hypo-methylated < 0.001 0.02
chrMT Mt-nd1 3.69 1.82 2.03 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd2 3.69 1.82 2.03 Hyper-methylated < 0.001 < 0.001
chrMT AY172581.24 3.69 1.82 2.03 Hyper-methylated < 0.001 < 0.001
chrMT Mt-co1 2.84 1.48 1.92 Hyper-methylated < 0.001 < 0.001
chr20 Rt1-ce10 1.75 1 1.76 Hyper-methylated 0.01 0.21
chr5 Tipinl1 12.83 8.55 1.5 Hyper-methylated < 0.001 0.11
chr15 Dlg5 1.03 0.69 1.5 Hyper-methylated 0.04 0.53
chr12 AABR07034833.1 14.75 11.11 1.33 Hyper-methylated 0.02 0.33
chrMT Mt-co3 0.41 0.31 1.32 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd4l 0.41 0.31 1.32 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd4 0.41 0.31 1.32 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd5 0.41 0.31 1.32 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd3 0.41 0.31 1.32 Hyper-methylated < 0.001 < 0.001
chrMT Mt-atp6 0.41 0.31 1.32 Hyper-methylated < 0.001 < 0.001
chrMT Mt-co2 0.37 0.31 1.22 Hyper-methylated < 0.001 < 0.001
chrMT Mt-atp8 0.52 0.45 1.16 Hyper-methylated < 0.001 < 0.001
chrMT Mt-cyb 0.36 0.32 1.13 Hyper-methylated < 0.001 < 0.001
chrMT Mt-nd6 0.36 0.32 1.13 Hyper-methylated < 0.001 < 0.001
chr4 Htr5a 4.43 5.77 0.77 Hypo-methylated < 0.001 0.23
chr18 Vom2r75 4.01 5.88 0.68 Hypo-methylated 0.05 0.55
chr5 AABR07046628.1 1.09 2.23 0.49 Hypo-methylated 0.02 0.44
chr19 LOC679149 1.64 3.64 0.45 Hypo-methylated 0.02 0.41
LPE: Early-life low-protein group, LPF: Fetal low-protein group, CON: Control group
Volume 1 Issue 2 (2022) 20 https://doi.org/10.36922/gpd.v1i2.169