Page 81 - TD-4-2
P. 81
Tumor Discovery DRGs in HCC prognosis and immunity
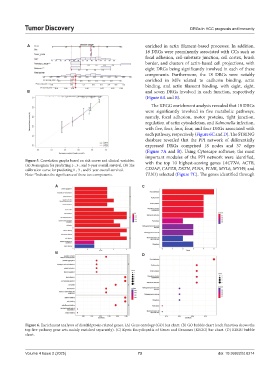
A enriched in actin filament-based processes. In addition,
18 DRGs were prominently associated with CCs such as
focal adhesion, cell-substrate junction, cell cortex, brush
border, and clusters of actin-based cell projections, with
eight DRGs being significantly involved in each of these
components. Furthermore, the 18 DRGs were notably
enriched in MFs related to cadherin binding, actin
binding, and actin filament binding, with eight, eight,
B and seven DRGs involved in each function, respectively
(Figure 6A and B).
The KEGG enrichment analysis revealed that 18 DRGs
were significantly involved in five metabolic pathways,
namely, focal adhesion, motor proteins, tight junction,
regulation of actin cytoskeleton, and Salmonella infection,
with five, four, four, four, and four DRGs associated with
each pathway, respectively (Figure 6C and D). The STRING
database revealed that the PPI network of differentially
expressed DRGs comprised 18 nodes and 37 edges
(Figure 7A and B). Using Cytoscape software, the most
important modules of the PPI network were identified,
Figure 5. Correlation graphs based on risk scores and clinical variables. with the top 10 highest-scoring genes (ACTN4, ACTB,
(A) Nomogram for predicting 1-, 3-, and 5-year overall survival. (B) The
calibration curve for predicting 1-, 3-, and 5-year overall survival. CD2AP, CAPZB, DSTN, FLNA, FLNB, MYL6, MYH9, and
Note: *Indicates the significance of these two components. TLN1) selected (Figure 7C). The genes identified through
A C
B D
Figure 6. Enrichment analyses of disulfidptosis-related genes. (A) Gene ontology (GO) bar chart. (B) GO bubble chart (each function shows the
top five pathway gene sets mainly enriched separately). (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) bar chart. (D) KEGG bubble
chart.
Volume 4 Issue 2 (2025) 73 doi: 10.36922/td.8214