Page 89 - AIH-1-2
P. 89
Artificial Intelligence in Health Combating XDR-bacteria as we approach 2050
Table 2. (Continued)
Accession number Size (kb) Multidrug‑resistant marker genes: Drug transporters GenBank (year) Pathogenic bacterial name
and antibiotic‑inactivating enzymes
GU585907 79 aadA2, aphA2, aadA1*, strA, strB, and blaVIM1 2010 K. pneumoniae
KF954759 73 blaKPC3, strB, aac (6’), and chrB 2014 K. pneumoniae
KJ460501 62 blaCTX-M 2014 S. sonnei
NZ_CP008901 52 Dhfr, blaKPC-2 2015 E. cloacae
AY046276 51 aadA1, blaOXA-2, sul1, tetA, and ABC 2012 S. enterica
KT225462 50 mphE, sul1, blaDHA-1, qbrB, strA, and strB 2015 K. pneumoniae
AB61665 47 blaIMP2, aacA4, aadA2, tetA, blaCTX-M, and sul1 2012 K. pneumoniae
KJ541071 44 sul1, blaOXA-2, aadA/B, blaTEM, catA1, and blaGES-5 2014 E. coli
JX104759 42 blaKPC-2 and ABC 2013 K. pneumoniae
KC354802 41 aacA4, aadA1, blaOXA-9, and blaTEM-1 2013 K. pneumoniae
NC_021087 26 blaGIM-1, aacA4, aadA1, blaOXA-2, and sul1 2015 E. cloacae
JN215524 24 Dhfr, cmlA, blaOXA10, aadA1, qnrB, blaDHA1, and sul1 2012 C. freundii
NG_035843 15 blaOXA-30, catB3, arr-3, sul1, qnr, and blaDHA-1 2014 E. coli
NG_041456 2.5 blaKPC-2 2014 P. aeruginosa
JN677524 1.9 Ble and blaNDM-1 2016 K. pneumonia
Abbreviations: P. aeruginosa: Pseudomonas aeruginosa; K. oxytoca: Klebsiella oxytoca; K. pneumonia: Klebsiella pneumonia; E. coli: Escherichia coli;
S. enterica: Salmonella enterica; C. freundii: Citrobacter freundii; P. stuartii: Providencia stuartii; E. aerogenes: Klebsiella aerogenes;
E. cloacae: Enterobacter cloacae; S. marcescens: Serratia marcescens; S. sonnei: Shigella sonnei.
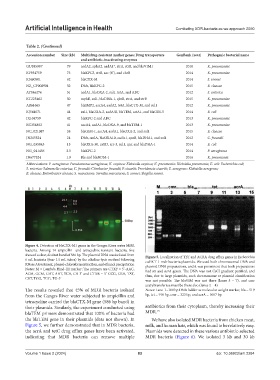
Figure 4. Detection of blaCTX-M1 genes in the Ganges River water MDR
bacteria. Among 14 ampicillin- and tetracycline-resistant bacteria, five
showed a clear, distinct band of 586 bp. The plasmid DNA was isolated from Figure 5. Localization of TET and ACRA drug efflux genes in Escherichia
6 mL bacteria (four 1.5 mL tubes) by the alkaline-lysis method following coli KT-1_mdr bacteria plasmids. We used both chromosomal DNA and
RNase A treatment, phenol-chloroform extraction, and ethanol precipitation plasmid DNA preparations, and it was prominent that both preparations
Notes: M = Lambda Hind-III marker; The primers are CTXF = 5’-AAC, had tet and acrA genes. The DNA was not CsCl gradient purified, and
ACM, GCM, GAT, AAT, TCA, CA-3’ and CTXR = 5’-CCG, CRA, TAT, thus, due to large plasmids, such chromosome or plasmid classification
CRT, TGG, TGG, TG-3’
was not possible. The blaVIM was not there (lanes 5 – 7), and cmr
acetyltransferase must be there also (lanes 2 – 4)
The results revealed that 45% of MDR bacteria isolated Notes: Lane-1=100 bp DNA ladder as molecular weight marker, bla = 519
from the Ganges River water subjected to ampicillin and bp, tet = 910 bp, cmr = 323 bp, and acrA = 1007 bp.
tetracycline carried the blaCTX-M gene (586 bp band) in
their plasmids. Similarly, the experiment conducted using antibiotics from their cytoplasm, thereby increasing their
blaTEM primers demonstrated that 100% of bacteria had MDR. 15
the blaTEM gene in their plasmids (data not shown). In We have also isolated MDR bacteria from chicken meat,
Figure 5, we further demonstrated that in MDR bacteria, milk, and human hair, which was found to be relatively easy.
the acrA and tetC drug efflux genes have been activated, Plasmids were detected in these various antibiotic-selected
indicating that MDR bacteria can remove multiple MDR bacteria (Figure 6). We isolated 3 kb and 30 kb
Volume 1 Issue 2 (2024) 83 doi: 10.36922/aih.2284