Page 94 - AIH-1-2
P. 94
Artificial Intelligence in Health Combating XDR-bacteria as we approach 2050
3.6. Target identification of CU1 as RNA polymerase which destroys gram-positive bacteria. 26,27 It disrupts
We further investigated the molecular target of CU1 bacterial membrane function and affects DNA and protein
and identified its inhibition of E. coli RNA polymerase structure. Cyclic peptide antibiotics are good and more
activity. Among the 11 phytochemicals tested, only CU1 resistant to inactivation by MDR enzymes. Research
exhibited inhibition comparable to the potent bacterial is ongoing for the development of salivaricins, nisins,
RNA polymerase inhibitor known as rifampicin. As and related lantibiotics for combating XDR tuberculosis
16
rifampicin is a potent anti-TB drug, we tested the efficacy (XDR-TB). AI is important in understanding the 3D
of the CU1 drug on Mycobacterium tuberculosis RNA structural interaction between drugs and target proteins.
polymerase, revealing dose-dependent inhibition with Computer-assisted modification of drugs generated a good
increasing concentrations of CU1 (Figure 20). However, antibiotic with target specificity. For example, 3D crystal
16
in the DNA polymerase assay, there was no inhibition of structure simulation clearly predicted the rifampicin drug’s
E. coli DNA polymerase by CU1 (Figure 21). CU1 has no specificity for the M. tuberculosis RNA polymerases and
potential inhibitory effects on the growth of mammalian was quite different from other similar RNA polymerases
cells in culture, revealing that CU1 plays no role in human of E. coli, S. aureus, and K. pneumoniae. Thus, rifampicin
RNA polymerase activity (data not shown). is a first-line drug despite the emergence of rpoB gene
(RNA polymerase beta-subunit) mutations that confer
3.7. Lantibiotics research has gained momentum resistance. However, other TB-specific drugs such as
with AI-guided and computer-assisted 3D graphics
of drug-enzyme interaction
The gramicidin peptide antibiotic has been long recognized
to cure skin infections. It is produced by Bacillus brevis,
Figure 22. Complex cyclic structures of salivaricin-B lantibiotic that are
effective against MDR bacteria. This peptide antibiotic was isolated from
the oral bacteria Streptococcus salivarious, which contained many large
plasmids with 10 salivaricin lantibiotic synthesizing genes
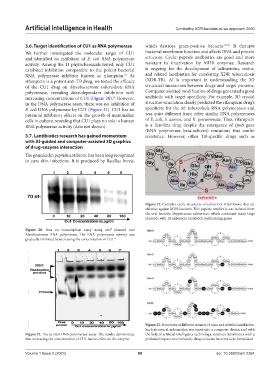
Figure 20. Run on transcription assay using sinP plasmid and
Mycobacterium RNA polymerase. The RNA polymerase activity was
gradually inhibited by increasing the concentration of CU1 15
Figure 23. Structures of different isomers of nisin and subtilin lantibiotics.
Such structural information was input into a computer device, and with
Figure 21. The in vitro DNA polymerase assay. The results demonstrate the help of artificial intelligence technology, synthetic lantibiotics with a
that increasing the concentration of CU1 has no effect on the enzyme profound impact on extensively drug-resistant bacteria were formulated
Volume 1 Issue 2 (2024) 88 doi: 10.36922/aih.2284