Page 92 - AIH-1-2
P. 92
Artificial Intelligence in Health Combating XDR-bacteria as we approach 2050
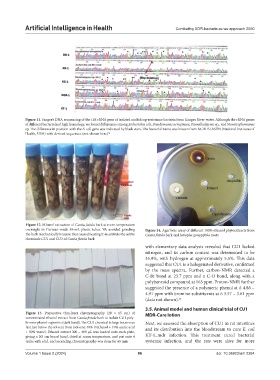
Figure 11. Sanger’s DNA sequencing of the 16S rRNA gene of isolated multidrug-resistance bacteria from Ganges River water. Although the rRNA genes
of different bacteria had high homology, we found differences among Escherichia coli, Pseudomonas aeruginosa, Paenalkaligenes sp., and Stenotrophomonas
sp. The difference in position with the E. coli gene was indicated by black stars. The bacterial name was known from NCBI BLASTN (National Institutes of
Health, USA) with derived sequences (not shown here) 14
Figure 12. Ethanol extraction of Cassia fistula bark at room temperature
overnight in German-made 50-mL plastic tubes. We avoided grinding Figure 14. Agar hole assay of different 100% ethanol phytoextracts from
the bark mechanically because that caused heating to inactivate the active Cassia fistula bark and Jatropha gossypifolia roots
chemicals CU1 and CU3 of Cassia fistula bark
with elementary data analysis revealed that CU1 lacked
nitrogen, and its carbon content was determined to be
35.9%, with hydrogen at approximately 5.5%. This data
suggested that CU1 is a halogenated derivative, confirmed
by the mass spectra. Further, carbon-NMR detected a
C-Br bond at 23.7 ppm and a C-O bond, along with a
polybenzoid compound at 165 ppm. Proton-NMR further
suggested the presence of a polymeric phenol at ό 4.86 –
4.91 ppm with bromine substituents at ό 3.57 – 3.61 ppm
(data not shown). 16
3.5. Animal model and human clinical trial of CU1
Figure 13. Preparative thin-layer chromatography (20 × 15 cm) of MDR-Cure lotion
concentrated ethanol extract from Cassia fistula bark to isolate CU1 poly-
bromo-phenol-saponins (dark band). The CU1 chemical is large but moves Next, we assessed the absorption of CU1 in rat intestines
fast just below the solvent front (solvent: 40% methanol + 10% acetic acid and its distribution into the bloodstream to cure E. coli
+ 50% water). Ethanol extract 300 – 400 µL was loaded onto each plate,
giving a 0.5 cm broad band, dried at room temperature, and put onto 4 KT-1_mdr infection. This treatment cured bacterial
tanks with a lid, and ascending chromatography was done for 65 min systemic infection, and the rats were alive for more
Volume 1 Issue 2 (2024) 86 doi: 10.36922/aih.2284