Page 91 - GPD-2-1
P. 91
Gene & Protein in Disease A pan-cancer analysis of HMGB1
A C
D
B E F
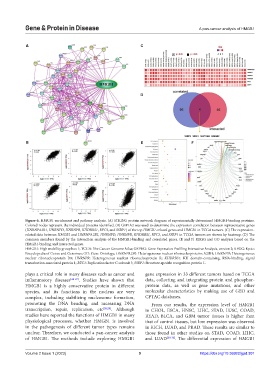
Figure 6. HMGB1 enrichment and pathway analysis. (A) STRING protein network diagram of experimentally determined HMGB1-binding proteins.
Colored nodes represent the individual proteins identified. (B) GEPIA2 was used to determine the expression correlation between representative genes
(HNRNPA2B1, HNRNPD, HNRNPR, KHDRBS1, RFC3, and SSRP1) of the top HMGB1-related genes and HMGB1 in TCGA tumors. (C) The expression-
related data between HMGB1 and HNRNPA2B1, HNRNPD, HNRNPR, KHDRBS1, RFC3, and SSRP1 in TCGA tumors are shown by heatmap. (D) The
common members found by the interaction analysis of the HMGB1-binding and correlated genes. (E and F) KEGG and GO analyses based on the
HMGB1-binding and interacted genes.
HMGB1: High mobility group box 1; TCGA: The Cancer Genome Atlas; GEPIA2: Gene Expression Profiling Interactive Analysis, version 2; KEGG: Kyoto
Encyclopedia of Genes and Genomes; GO: Gene Ontology; HNRNPA2B1: Heterogeneous nuclear ribonucleoproteins A2/B1; HNRNPD: Heterogeneous
nuclear ribonucleoprotein D0; HNRNPR: Heterogeneous nuclear ribonucleoprotein R; KHDRBS1: KH domain-containing, RNA-binding, signal
transduction-associated protein 1; RFC3: Replication factor C subunit 3; SSRP1: Structure-specific recognition protein 1.
plays a critical role in many diseases such as cancer and gene expression in 33 different tumors based on TCGA
inflammatory diseases [24-27] . Studies have shown that data, collecting and integrating protein and phosphor-
HMGB1 is a highly conservative protein in different protein data, as well as gene mutations, and other
species, and its functions in the nucleus are very molecular characteristics by making use of GEO and
complex, including stabilizing nucleosome formation, CPTAC databases.
promoting the DNA bending, and increasing DNA From our results, the expression level of HMGB1
transcription, repair, replication, etc [28,29] . Although in CHOL, ESCA, HNSC, LIHC, STAD, LUSC, COAD,
studies have reported the functions of HMGB1 in many READ, BLCA, and GBM tumor tissues is higher than
physiological processes, whether HMGB1 is involved that of control tissues, but low expression was observed
in the pathogenesis of different tumor types remains in KICH, LUAD, and PRAD. These results are similar to
unclear. Therefore, we conducted a pan-cancer analysis those found in other studies on STAD, COAD, LIHC,
of HMGB1. The methods include exploring HMGB1 and LUAD [30-33] . The differential expression of HMGB1
Volume 2 Issue 1 (2023) 9 https://doi.org/10.36922/gpd.301