Page 82 - TD-3-1
P. 82
Tumor Discovery Missense mutations in CXCR1: Impact on stability and function
A
B
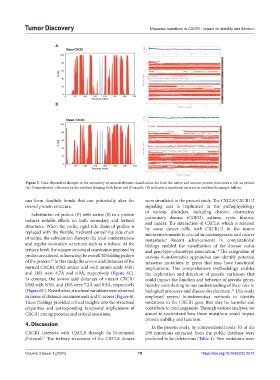
Figure 5. Time-dependent changes in the secondary structural element classification for both the native and mutant protein structures a 100-ns period.
(A) Demonstrated a decrease in the residues forming both loops and β-strands. (B) Indicates a significant increase in residues forming α-helices.
can form disulfide bonds that can potentially alter the were simulated in the present study. The CXCL8-CXCR1/2
overall protein structure. signaling axis is implicated in the pathophysiology
of various disorders, including chronic obstructive
Substitution of proline (P) with serine (S) in a protein pulmonary disease (COPD), asthma, cystic fibrosis,
induces notable effects on both secondary and tertiary and cancer. The interaction of CXCL8, which is released
structures. When the cyclic, rigid side chain of proline is by some cancer cells, with CXCR1/2 in the tumor
replaced with the flexible, hydroxyl-containing side chain microenvironment is crucial in carcinogenesis and cancer
of serine, the substitution disrupts the local conformations metastasis. Recent advancements in computational
6
and regular secondary structures such as α-helices. At the biology enabled the visualization of the disease status
tertiary level, the unique structural constraints imposed by and genotype–phenotype association. The integration of
70
proline are altered, influencing the overall 3D folding pattern various bioinformatics approaches can identify potential
of the protein. In this study, the amino acid distances of the missense mutations in genes that may have functional
69
normal CXCR1 P302 amino acid with amino acids N301 implications. This comprehensive methodology enables
and I303 were 6.7Å and 6.9Å, respectively (Figure 6C). the exploration and detection of genetic variations that
In contrast, the amino acid distances of mutant CXCR1 could impact the function and behavior of specific genes,
S302 with N301 and I303 were 7.2Å and 8.9Å, respectively thereby contributing to our understanding of their role in
(Figure 6C). Nonetheless, structural variations were observed biological processes and disease development. This study
71
in terms of distance measurements and C-scores (Figure 6). employed several bioinformatics methods to identify
These findings provided critical insights into the structural mutations in the CXCR1 gene that may be harmful and
disparities and corresponding functional implications of contribute to carcinogenesis. Through various analyses, we
CXCR1 among proteins and critical mutations. aimed to understand how these mutations could impact
protein stability and function.
4. Discussion
In the present study, by computational tools, 53 of the
CXCR1 interacts with CXCL8 through its N-terminal 299 mutations extracted from the public database were
β-strand. The tertiary structures of the CXCL8 dimers predicted to be deleterious (Table 1). Five mutations were
17
Volume 3 Issue 1 (2024) 18 https://doi.org/10.36922/td.2512